Distance Measurements by EPR Spectrosopy
Electron paramagnetic resonance (EPR) spectroscopy measures changes in the energy levels of paramagnets upon placement in a magnetic field. The technique is capable of measuring the types of centre and investigating other properties such as changes in environment and dynamics. Our lab most often uses the technique to measure nanometre (in the region of 0.000000001 m) distances between pairs of paramagnetic centres. Typically the paramagnet is an atom or group of atoms containing one, but sometimes more, unpaired electron spins and these are frequently called radicals. EPR spectroscopy is capable of measuring this scale with a high degree of accuracy and this makes it an important biophysical tool for investigating biomolecule, and particularly, protein structure.

So how do we use EPR spectroscopy to measure nanometre scale distances? Imagine a pair of bar magnets, one in each hand. As you move your hands towards each other the magnets either attract or repel each other. As you continue the force gets stronger. The magnetic field is sensed by each magnet in a distance (and orientation!) dependent manner. Our unpaired electrons are a little bit like bar magnets in so much as they have a magnetic, called a dipolar, field associated with them (see the picture on the right, the electrons are represented by red arrows, one of them has dipolar field lines added and r is the distance between them). They can interact with each other to change their energy levels. Of course this is at the quantum level and this is only a way to imagine it, rather than an entirely accurate description - for example there is also the Heisenberg Exchange Interaction to consider.
At very short distances, for example if the unpaired electrons are all on a single atom as for certain transition metal elements/ions, the force is very large and this is called the zero-field splitting. As the electrons become more separated in space the force decreases and at somepoint it is valid to estimate that the dipole-dipole coupling between the electrons depends on the inverse cube of the distance between them. Therefore, by somehow measuring the dipole-dipole coupling we can measure distances. This exact distance range will depend upon the paramagnets themselves. For the popular nitroxyl spin labels, where the single unpaired electron is primarily shared between a nitrogen and an oxygen atom, this distance range starts at 1.5 nm and has so far been measured up to nearly 13 nm (this latter result was measured in St Andrews by Dr Hassane El-Mkami and Dr David G. Norman in 2015).
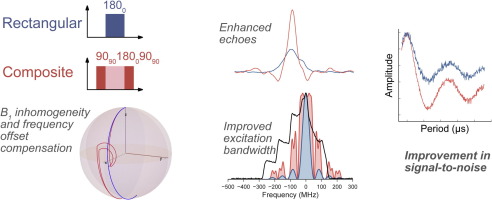 There are lots of EPR spectroscopic methods to measure the dipolar coupling (and therefore distances) but the one we use principally, and probably the most popular, has two names: PELDOR (pulsed electron electron resonance) and DEER (double electron electron resonance). The DOUBLE refers to the fact that the techniques use pulses of microwaves at two different frequencies to perform the experiment. We are interested in developing these methods to e.g. increase sensitivity or scope.
There are lots of EPR spectroscopic methods to measure the dipolar coupling (and therefore distances) but the one we use principally, and probably the most popular, has two names: PELDOR (pulsed electron electron resonance) and DEER (double electron electron resonance). The DOUBLE refers to the fact that the techniques use pulses of microwaves at two different frequencies to perform the experiment. We are interested in developing these methods to e.g. increase sensitivity or scope.
We measure frozen solutions of proteins. The solutions can contain a wide variety of components such as salts, detergents or membranes. The samples need to be frozen both to improve how long the paramagnets can give signals for and because the actual dipolar talk between the paramagnets can average to nothing if everything tumbles around in a liquid. Conditions are currently being pioneered to mean that measurements at normal temperatures may become possible. The sensitivity of the experiment until quite recently was at a level where we’d want at least 0.1 mM of the paramagnet and about 0.06 ml sample. The sensitivity is increasing though and we often now measure 0.01 mM.
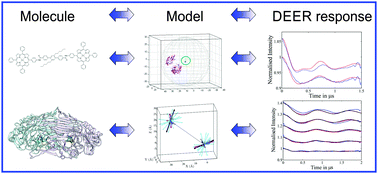
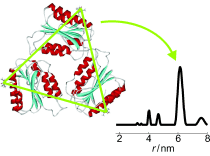 The DEER experiment gives a result in time and this is analysed by packages such as DeerAnalysis to give the distances and distance distributions. When the experiment is used to measure paramagnets which are quite well defined in space with respect to each other the distances can be very accurate, down to 0.05 of a nanometre.
The DEER experiment gives a result in time and this is analysed by packages such as DeerAnalysis to give the distances and distance distributions. When the experiment is used to measure paramagnets which are quite well defined in space with respect to each other the distances can be very accurate, down to 0.05 of a nanometre.
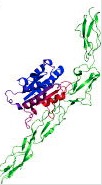 The distances and any angles that we obtain are used to refine structures of biomolecules or other materials. For example we can use the distances as restraints for modelling protein-protein docking.
The distances and any angles that we obtain are used to refine structures of biomolecules or other materials. For example we can use the distances as restraints for modelling protein-protein docking.
Spin Labels

 Some proteins contain suitable paramagnetic centres for the DEER distance measurements. Examples of suitable centres include semiquinones and copper. However, often proteins do not contain suitable sites naturally or more distances are needed to investigate their structure or conformational changes. Here we can add on spin labels. These are small molecules which contain a paramagnetic centre. Often these labels contain the nitroxyl radical, i.e. NO. Sometimes they may be more exotic and contain gadolinium or stable carbon radicals. Others have bound copper to specific amino acid sequences in proteins and used these coppers.
Some proteins contain suitable paramagnetic centres for the DEER distance measurements. Examples of suitable centres include semiquinones and copper. However, often proteins do not contain suitable sites naturally or more distances are needed to investigate their structure or conformational changes. Here we can add on spin labels. These are small molecules which contain a paramagnetic centre. Often these labels contain the nitroxyl radical, i.e. NO. Sometimes they may be more exotic and contain gadolinium or stable carbon radicals. Others have bound copper to specific amino acid sequences in proteins and used these coppers.
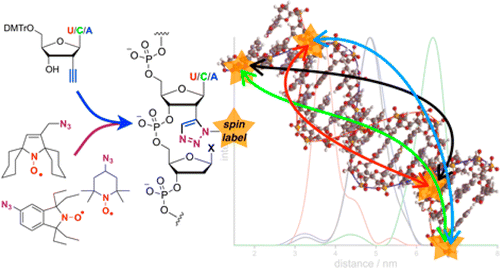
 Nitroxide spin labels have been used for 50 years or so for looking at the environment and movement of proteins (they are good at looking at movement on a nanosecond timescale) and as such they have been developed for easy attachment to proteins. Normally this attachment is via a cysteine amino acid which has a convenient thiol (SH) handle. One aim of our research is to make this attachment possible for proteins which contain multiple cysteines which would otherwise be accidentally labelled. Another aim of our work is to enable base-independent labelling of nucleic acids. We are also interested in exploring different kinds of spin labels and are actively looking at Gd.
Nitroxide spin labels have been used for 50 years or so for looking at the environment and movement of proteins (they are good at looking at movement on a nanosecond timescale) and as such they have been developed for easy attachment to proteins. Normally this attachment is via a cysteine amino acid which has a convenient thiol (SH) handle. One aim of our research is to make this attachment possible for proteins which contain multiple cysteines which would otherwise be accidentally labelled. Another aim of our work is to enable base-independent labelling of nucleic acids. We are also interested in exploring different kinds of spin labels and are actively looking at Gd.
Proteins
Proteins are polymers of amino acids. This means that a protein is made up from dozens or thousands of amino acids attached together. Each amino acid (of which there are about 20) has a different chemical group on it. Importantly the protein has a specific way of folding up the long chain of amino acids. The amino acids present and the way the protein is folded up determines the properties of the protein (this is a rather simplified picture, proteins are often modified after they are made by phosphorylation or glycosylation for example). Proteins perform most of the tasks in the body and they do this through interactions with each other and often quite subtle changes in their shape. Understanding more about the shape of the proteins can offer key insight into what they do and how they do it. This is what we want to know. This information is ultimately not only great to know but also feeds into work on drug discovery and protein mimicry.
Nucleic acids
Nucleic acids are polymers of nucleotides and these then fold into structures, rather like proteins but with fewer building blocks: there are fewer nucleotide types than amino acids. For example deoxyribose nucleic acid (DNA) forms pairings between nucleotides: G:C and A:T called Watson-Crick pairs and classically these form a double helix. Nucleic acids store the genetic material of an organism but are also used to translate that material into proteins or are sometimes used directly for functions.