The importance of co-transcriptional RNA folding: Cellular functions of RNA sequences are intrinsically associated to their structure and folding. Most folding studies today are carried out in full-length RNA molecules after transcription is finished. However, RNA has great propensity to fold while it is being transcribed and many important RNA molecules perform their biological function exclusively at transcriptional level, as in the case of riboswitches. RNA function can also become more efficient during the time frame of the transcription process, as it has been found in the Tetrahymena group I intron, RNase P RNA and self-cleaving small ribozymes such as HDV, hairpin and hammerhead ribozymes. The recent discovery of ~106 human RNA transcripts longer than 150 nt,and that co-transcriptional formation for some of these structures may be intrinsic to their function in processes such as chromatin remodelling of RNA methylation, emphasise the important biological role of co-transcriptional folding. These findings and the growing number of processes that are known to occur transcriptionally, highlight the need to develop novel tools that allow in-situ monitoring of RNA folding and function while it is being transcribed. One of the main obstacles to progress in this area remains in the inability to observe, in real-time, RNA folding events while RNA is being transcribed and therefore, to correlate changes in the structure of the nascent RNA with its biological function and its effect on the progress of the transcription machinery.
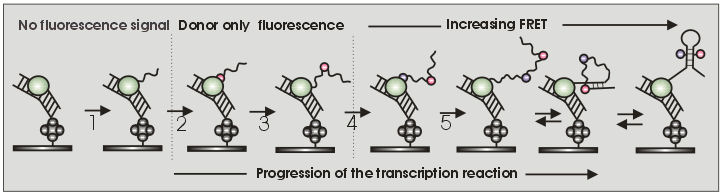 |
Schematics of the single-molecule stepwise transcription FRET assay. Immobilization of the RNAP-DNA complex on the glass surface; (1) Transcription progress until formation of the TEC or a user-defined position; (2) Single-step nucleotide incorporation of a donor or acceptor dye and subsequent detection of its corresponding fluorescence emission; (3) Transcription continues until a predefined second position; (4) Single-step incorporation of the second dye (donor or acceptor) and appearance of the FRET signal (low FRET state); (5) transcription continues by incorporation of the remaining nucleotides and transient folding intermediates emerge as the transcription progress (6) and (7) the full-length nascent RNA achieves its most stable conformation (e.g., higher FRET state) |
We aim to develop a new experimental framework by simultaneous combination of single-molecule fluorescence, microfluidics and stepwise transcription that will allow an unprecedented characterization of nascent RNA and its interactions with the rest of elements involved in the transcription scaffold. The single-molecule transcription technique will allow for the first time to monitor the folding of any RNA molecule in real-time during the transcriptional process at the nucleotide resolution.According to this methodology and once the FRET dyes have been incorporated into the nascent RNA following the stepwise assay, two distinct readout regimes can be followed: (i) static regime or structure snapshot, where the transcription is halted at a certain position and the folding dynamics of that particular transcribed portion is examined for a period of time by recording the fluctuations on the FRET signal and (ii) real-time regime where after the donor and acceptor dyes have been incorporated, the transcription is allowed to run in normal mode. Thus, we can monitor in a single trace the progression of the transcription reaction and its effect on the folding dynamics of that particular RNA region following the changes in the donor and acceptor signals. This project is in collaboration with Prof. Daniel Lafontaine: Lafontaine's Lab