Thermostability

Citrate synthase
The PDB code links will take you to the PDB entry with links to the publication
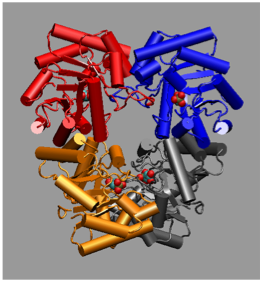
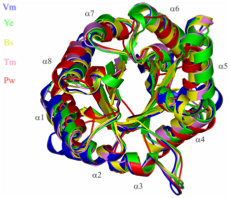
A comparative structural study was undertaken to establish the basis of heat and cold tolerance. Citrate synthase was used as a model enzyme, and structures were determined from organisms spanning the temperature rang eat which life is found: 0C to >100C. Factors such as networks of ion paris (shown below) and removal of internal cavities correlated with increasing temperature. This study was in collaboration with Mike Danson & David Hough at the University of Bath.

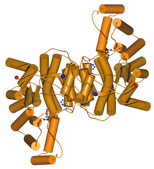

Antartic bacterium (0C) T. acidophilum (55C) S. solfataricus (85C) P. furiosus (100C)


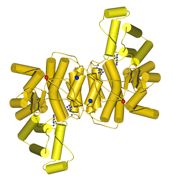
Triosephosphate isomerase
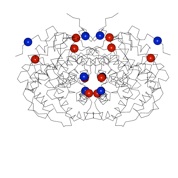

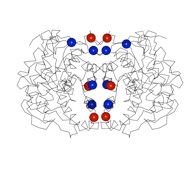

Most TIMs are dimers, but in thermophilic archaea they are tetramers. The P. furiosus TIM has a smaller monomer, and achieves thermostability through shorter loops and a predominantly hydrophobic interaction of two classical dimers to form the tetramer. This study was in collaboration with Mike Danson & David Hough at the University of Bath.