Viral sialidases

The PDB code links will take you to the PDB entry with links to the publication
Sialidases (neuraminidases) remove sialic acid, a 9-carbon carbohydrate that decorates cell surfaces as the terminal sugar of various glycoconjugates. The influenza virus neuraminidase is the target of two drugs used in the treatment of influenza: Tamiflu and Relenza. We are studying sialidases from viral, bacterial and mammalian sources, with a view to structure-based drug discovery. This work is a collaboration with Dr Allen Portner, St Jude Children’s Research Hospital, Memphis.
Paramyxovirus hemagglutinin-neuraminidase
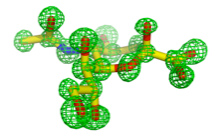
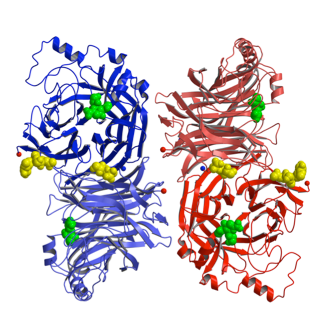
The HN glycoprotein of paramyxoviruses has three functions: it binds to sialic acid receptor, triggers the fusion (F) protein and removes sialic acids from progeny virus particles. Our structure of the first HN from Newcastle disease virus (NDV) revealed an unusual tetrameric structure formed from a dimer of dimers. A single sialic acid binding site can switch between a binding site and a catalytic site. Variations in the dimer led us to propose a mechanism for HN triggering the fusion protein (movie below). NDV HN, and possibly hPIV1-HN have a second sialic acid binding site (yellow below) at the dimer interface.

With Biocryst Pharmaceuticals, we have developed specific inhibitors of HN (BCX2798) that protect mice from a lethal parainfluenza virus.

This work is a collaboration with Dr Allen Portner, St Jude Children’s Research Hospital, Memphis.
PDB codes: 1e8t (low pH, apo)




